Top synthetic biology news from academic labs in 2019
Oct 23, 2019
This week, we are bringing you some of the hottest research news in synthetic biology in 2019: what were the most exciting academic articles published this year by synthetic biology researchers, and why?On Monday, scientists reported in Nature a new DNA editing tool, prime editing, that can be a real search-and-replace function for DNA, and repair any of the 75,000 known mutations that cause inherited disease in humans. Unlike base editors, which can perform 25% of possible nucleotide substitutions, prime editors have the ability to change any nucleotide to any other nucleotide, according to GEN, and the technology makes fewer off-target edits when compared with CRISPR-Cas9.“You can think of prime editors as being like word processors capable of searching for target DNA sequences and precisely replacing them with edited DNA sequences,” said David Liu of the Broad Institute for The Guardian. “Potential impacts include being able to directly correct a much larger fraction of the mutations that cause genetic diseases, and being able to introduce DNA changes into crops that result in healthier or more sustainable foods,” Liu said.
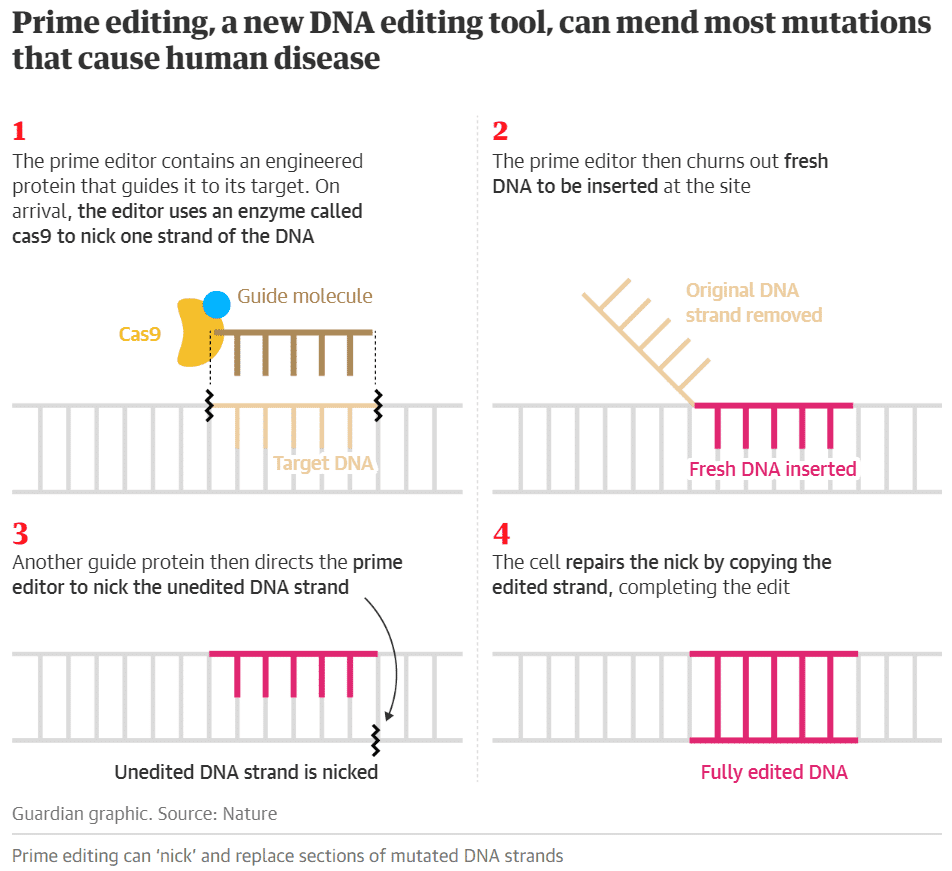
Image: The Guardian.The Broad Institute has applied for a patent on prime editing, and is already making it freely available to academic and non-profit researchers for non-commercial uses. It has also given an exclusive license for the commercial development of human therapeutics to Prime Medicine, a new company co-founded by Liu, according to STAT News.What about other research projects this year? Check out the comments from some academics:Dr. Tom Ellis, Professor of Synthetic Genome Engineering at Imperial College London, said “Jason Chin’s group at The MRC Laboratory of Molecular Biology showed they could synthesize the whole E. coli genome with recoding and also that they could rearrange the layout of the genome.” “They created a strain that isn’t using three of the codons that the rest of nature does,” he told STAT.Niko McCarty, PhD student in Bioengineering at Caltech: “CRISPR Multiplexing is fully coming of age, with 25 CRISPR RNAs expressed from a single array in mammalian cells.” Researchers at ETH Zurich have refined the CRISPR-Cas technology: they reported in August that it is now possible to modify dozens, if not hundreds, of genes in a cell simultaneously.Isaac Larkin, PhD candidate at Northwestern, said: “Some of my favorites were Zhang lab's CRISPR transposon paper, because of its potential applications for efficient delivery and insertion of large genetic payloads; Baker lab's paper demonstrating that gamers playing FoldIt could design stable globular proteins from scratch; the implementation of an integral feedback controller genetic circuit in bacteria, because the more control theory gets applied to bioengineering the better and more reliable bioengineering will get.”Isaac also mentioned the Baker lab's work demonstrating the therapeutic potential of designed proteins and the Jewett lab's collaboration with LanzaTech that “showed that rapid cell-free prototyping could be used to optimize metabolic pathway performance, and that the improvements worked when transferring from a cloning workhorse like E. coli lysate into an industrial bacterial strain from a completely different branch of the tree of life.”Dr. Leonardo Rios, Lecturer at Edinburgh University, highlighted a study by MIT and Harvard that demonstrated how programmable CRISPR-responsive smart materials could open the door to novel tissue engineering, bioelectronic, and diagnostic applications. Another interesting study was by Columbia researchers, who developed a method that could prevent human-engineered proteins from spreading into the wild. “Useful for the big challenge of synbio containment,” according to Rios.Rios also mentioned Hachimoji DNA and RNA: Hoshika et al. added an additional four synthetic nucleotides to produce an eight-letter genetic code and generate so-called hachimoji DNA. “With more diversity in the genetic building blocks, scientists could potentially create RNA or DNA sequences that can do things better than the standard four letters, including functions beyond genetic storage,” according to Nature.Another great study this year was by Professor Sang Yup Lee of KAIST and his team, who developed a strain that demonstrated the highest efficiency in producing fatty acids and biodiesels ever reported.Rios also mentioned the study by UC Berkeley researchers who found a way to make the main chemicals in marijuana, THC and CBD, from yeast; one by Caltech researchers who used machine-learning-guided directed evolution for protein engineering; and a cloning method, COMPASS, that allows building tens to thousands of different plasmids in a single cloning reaction tube.Here are some of the most popular tweets and Facebook posts about research in the field this year:
October
https://twitter.com/SynBioBeta/status/1180630748350619648
September
https://twitter.com/SynBioBeta/status/1173031173972512768https://www.facebook.com/SynBioBeta/posts/1552669998207858
August
https://twitter.com/SynBioBeta/status/1166085417617301508https://www.facebook.com/SynBioBeta/posts/1512637672211091
July
https://twitter.com/SynBioBeta/status/1146835085787897859https://www.facebook.com/SynBioBeta/posts/1491416670999858
June
https://twitter.com/SynBioBeta/status/1138435721503674368https://www.facebook.com/SynBioBeta/posts/1479614085513450
May
https://twitter.com/SynBioBeta/status/1131296421972512769https://www.facebook.com/SynBioBeta/posts/1450126455128880
April
https://twitter.com/SynBioBeta/status/1118123387124109314https://www.facebook.com/SynBioBeta/posts/1429717593836433
March
https://twitter.com/SynBioBeta/status/1110497885811101701https://www.facebook.com/SynBioBeta/posts/1397113097096883
February
https://twitter.com/SynBioBeta/status/1098786960284573696https://www.facebook.com/SynBioBeta/posts/1394522884022571
January
https://twitter.com/SynBioBeta/status/1085929488658120708What do you think were the most exciting academic papers this year? Let us know in your comments below.

















