Novel Biopolymers: Creating a Fully Programmable Protein-Translation System
Jun 15, 2023
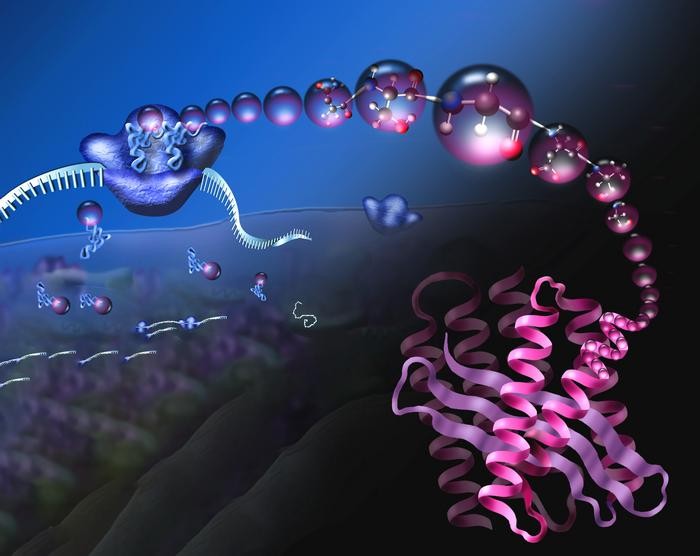
Ribosomes (blue, upper left) are nanomachines that read mRNA (coming in from left) to assemble a chain of amino acids (magenta balls) that folds into a compact 3D protein (lower right, pink). Image adapted from NSF (Wikipedia)
A cross-institutional group of chemists is aiming high with their newest endeavor: reengineering the internal workings of cells, specifically ribosomes, to create more complex polymer chains. This revolutionary work is being led by the University of California, Berkeley, as part of a $20 million research initiative. The researchers are making headway, as detailed in their latest publications, where they address key challenges, such as redesigning cells to provide ribosomes with alternative building blocks, predicting the most efficient building blocks, and altering ribosomes to use these new components in polymer creation.
The ultimate vision of the National Science Foundation Center for Genetically Encoded Materials (C-GEM) is to revolutionize the translation system, allowing it to be fully programmable. This would mean that introducing mRNA instructions into the cell with new building blocks would enable the ribosome to create an array of novel molecular chains. These new chains could be the foundation for innovative bio-materials, enzymes, or pharmaceuticals.
New research published in Nature Chemistry and ACS Central Science paves the way to transform synthetic cellular machinery to produce novel polymers, including bio-polymers and peptide macrocycles. The implications of these developments are vast, with potential applications that could be predetermined or completely unexpected.
C-GEM director Alanna Schepartz, PhD., explained, "C-GEM is working to biosynthesize molecules that have never before been made in a cell and are designed to have unique properties. The tools could be applied broadly by polymer chemists, medicinal chemists, and biomaterials scientists to generate bespoke materials with new functions." She highlighted an example of programming the ribosome to synthesize a polymer that merges the strengths of spider silk and nylon, a feat achievable with the technology being developed by C-GEM.
Jamie Cate, PhD., a UC Berkeley professor, highlighted the possibilities a programmable ribosome machine opens up. "We've had protein polymers evolving on the planet for billions of years, but we've been restricted in what those polymers are because the building blocks are the same 20 amino acids. If we can develop a system where you can actually apply evolution to these new polymers, then it's like a platform that anybody who has a creative idea can use to evolve a polymer to something they want."
A decade ago, this initiative seemed impossible. Now, C-GEM is pushing ahead to redefine what's possible in synthetic biology, thanks mainly to the relentless efforts of Schepartz. The team is working to supply the ribosome with alternative building blocks, an ambitious goal already showing signs of success, as detailed in a recent Nature Chemistry paper.
Schepartz added, "If we could help solve that problem by generating novel molecules whose functions encode unique modes of action, that would be an enormous contribution." However, this ambitious project must be accomplished within a living cell without disturbing the existing processes necessary for life, as Schepartz and Cate reiterated.
Other collaborators on this pioneering project include Matthew Francis from UC Berkeley, Scott Miller from Yale University, Abhishek Chatterjee from Boston College, Bhavana Shah, Zhonqi Zhang from Amgen Inc., and C-GEM managing director Sarah Smaga. Schepartz and Cate are faculty scientists at Lawrence Berkeley National Laboratory, with Schepartz also a member of the Chan Zuckerberg Biohub and the California Institute for Quantitative Biosciences (QB3). At the same time, Cate is a member of the Innovative Genomics Institute.