Move Over, CRISPR. Smaller, Smarter Gene-Editing System Found
A newly discovered RNA-guided gene-editing system, called TIGR, is smaller and more precise than CRISPR, making it a promising tool for gene therapy
Feb 27, 2025
[Canva]
For decades, the gene-editing revolution has been dominated by CRISPR—molecular scissors that can precisely slice DNA. But CRISPR has limitations. It’s big, hard to deliver, and restricted in where it can cut. Now, researchers at MIT’s McGovern Institute and the Broad Institute of MIT and Harvard have found something potentially better: a compact, modular, RNA-guided system that could change everything.
Meet TIGR—short for Tandem Interspaced Guide RNA. It’s small. It’s programmable. And unlike CRISPR, it doesn’t need special DNA sequences to work. That means scientists could, in theory, use it anywhere in the genome. The discovery, published in Science on February 27, 2025, could open the door to simpler, more effective gene-editing therapies.
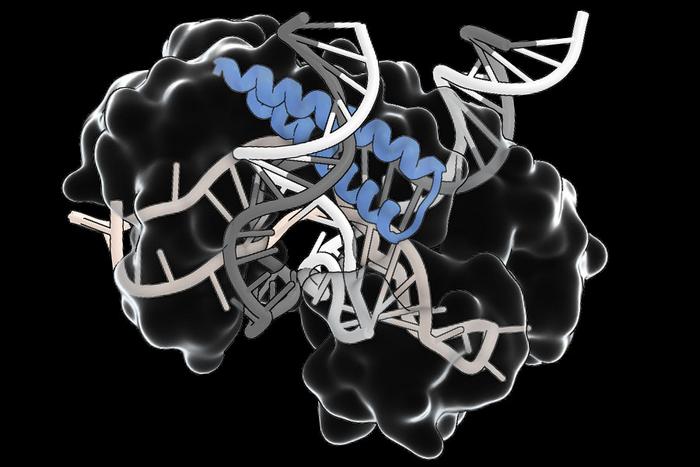
“This is a very versatile RNA-guided system with a lot of diverse functionalities,” says Feng Zhang, the MIT scientist behind some of the biggest breakthroughs in gene editing. Zhang’s team found TIGR lurking in an unexpected place: the genomes of viruses that infect bacteria. These systems use small RNAs to direct proteins, called Tas, to precise locations in DNA. Some Tas proteins can cut DNA directly. Others seem to recruit different molecular machines. That modularity could make TIGR a powerful addition to the gene-editing toolkit.
Mining Nature’s Genetic Treasure Trove
This isn’t the first time Zhang’s team has found gold by digging into microbial genetics. They’re the same group that turned bacterial CRISPR systems into the gene-editing tools now used around the world. But CRISPR has its downsides. The system depends on what are called protospacer adjacent motifs, or PAMs—short DNA sequences that act as landing pads. If a gene lacks the right PAM, CRISPR can’t touch it.
TIGR doesn’t have that problem.
To find it, Zhang’s team started with a simple question: Are there other RNA-guided systems out there? They zeroed in on a crucial feature of CRISPR’s Cas9 protein—the part that binds to guide RNA. Then, they scanned hundreds of millions of known and predicted protein structures, looking for anything similar. The first clue led them to a bacterial protein called IS110, which can also bind RNA. That led to more searches. The results kept piling up.
At a certain point, there was too much data to analyze with conventional evolutionary tools. So the team turned to artificial intelligence. A protein language model helped sort the results, grouping related systems together. One stood out: a family of proteins that, like CRISPR, were encoded alongside repetitive RNA sequences. These were the TIGR-Tas systems.
The team found more than 20,000 different Tas proteins, mostly in bacterial viruses. These proteins use short RNA guides to locate their targets, just like CRISPR. Some include a built-in DNA-cutting domain. Others seem to work in teams, binding to additional cellular machinery.
The Future of Gene Editing?
The real breakthrough here isn’t just that TIGR is RNA-guided. It’s that it’s tiny. On average, Tas proteins are a quarter of the size of CRISPR’s Cas9. That makes them far easier to deliver into human cells—one of the biggest challenges in gene therapy.
And then there’s the targeting issue. Because TIGR doesn’t need a PAM sequence, it could, in theory, reach any spot in the genome. “This means, theoretically, any site in the genome should be targetable,” says Rhiannon Macrae, a scientific advisor on the project.
TIGR may also be more precise. Unlike CRISPR, which uses a single guide RNA, some TIGR systems appear to rely on a “dual-guide” mechanism, interacting with both strands of DNA. That extra layer of specificity could help reduce off-target effects—one of the biggest concerns in gene editing today.
From Discovery to Application
Zhang’s team is already working on ways to refine TIGR for real-world use. They’ve mapped the molecular structure of one Tas protein that works in human cells, a crucial step toward engineering better versions. They’re also exploring whether these systems have deeper biological connections to human RNA-processing proteins—hints that nature may have already built similar mechanisms in our own cells.
If they’re right, TIGR could be more than just another gene-editing tool. It could be the next major leap in genetic medicine.
And if history is any guide, Zhang’s team is just getting started.


















