‘mEnrich-ing’ Microbiome Research with Groundbreaking Sequencing Technique
Mount Sinai researchers unveil mEnrich-seq, a revolutionary method promising unprecedented precision in microbiome research and potential breakthroughs in healthcare
Jan 4, 2024
AI Image Created Using DALL-E
Over the past decade, research into the human microbiome has uncovered some amazing interactions of these microbial communities in the development of disease, as well as maintaining good health. Yet, studying the microbiome—human, animal, plant, or otherwise—is challenging, to say the least. However, a groundbreaking advancement has emerged from investigators from the Icahn School of Medicine at Mount Sinai that might just be a game changer. The introduction of mEnrich-seq, detailed in a recent publication in Nature Methods, marks a pivotal shift in how scientists approach the study of microbiomes. These complex communities of microorganisms, integral to human health, can now be examined with unprecedented specificity and efficiency, paving the way for profound insights and medical breakthroughs.
Delving into the Microbial Labyrinth with mEnrich-seq
Microbiomes, the diverse universe of microbes within us, are key to our well-being. Their imbalance or reduced diversity can herald a multitude of diseases. Traditional microbiome research often zeroes in on specific bacteria types, akin to finding a needle in a haystack. This is where mEnrich-seq redefines the game.
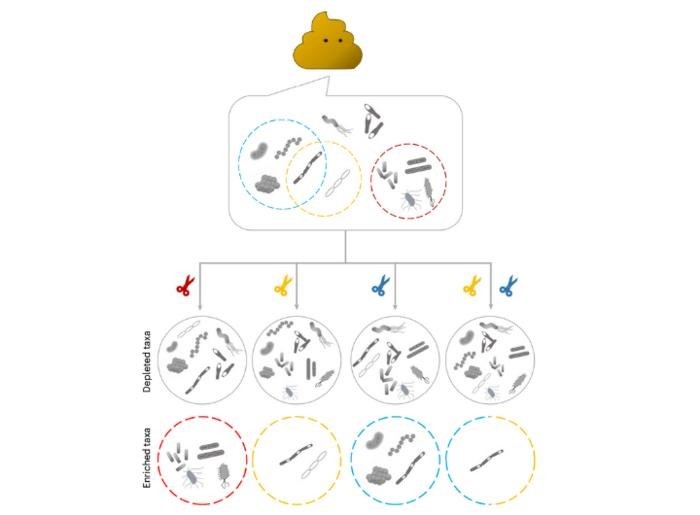
Gang Fang, PhD, Professor of Genetics and Genomic Sciences and the study's lead, illustrates this challenge: "Imagine you're a scientist who needs to study one particular type of bacteria in a complex environment. It's like trying to find a needle in a large haystack." mEnrich-seq, as he describes, is akin to a 'smart tweezer,’ adeptly extracting the sought-after 'needle.'
This technology transcends the previous need for culturing specific bacterial strains, a process often stymied by inefficiency and selectivity. mEnrich-seq, instead, directly retrieves targeted bacterial genomes from samples, revolutionizing the process.
It achieves this by leveraging unique “secret codes” in bacterial DNA, part of their native immune systems, allowing for precise differentiation and selection.
Redefining Research and Healthcare
The implications of mEnrich-seq are vast:
Cost-Effectiveness: This method promises a more budget-friendly avenue for extensive microbiome research.
Broad Applicability: Its versatility extends to a wide array of bacteria, enhancing both research and clinical applications.
Medical Advancements: mEnrich-seq's targeted approach could fast-track the development of novel diagnostics and treatments.
Dr. Fang highlights an exciting prospect: "One of the most exciting aspects of mEnrich-seq is its potential to uncover previously missed details, like antibiotic resistance genes that traditional sequencing methods couldn't detect due to a lack of sensitivity."
The study showcases mEnrich-seq's prowess in reconstructing pathogenic E. coli genomes from urine samples, a crucial step in analyzing antibiotic resistance. Another application involved the elusive Akkermansia muciniphila, a bacterium with potential benefits against obesity, diabetes, and cancer immunotherapy.
Envisioning the Future of mEnrich-seq
The team at Mount Sinai is not resting on their laurels. They are determined to enhance mEnrich-seq's efficiency and broaden its application scope. Collaborations with clinicians are underway to validate its effectiveness in practical settings.
Dr. Fang envisions a bright future for mEnrich-seq, foreseeing its role as a sensitive and versatile instrument in the advancing arena of microbiome studies and clinical applications, a testament to the relentless pursuit of scientific innovation and its potential to transform health care.