Evo: The AI That's Decoding Life's Genetic Blueprint
Trained on 2.7 million microbial genomes, Evo leverages deep learning to predict how small DNA changes affect organism fitness and can generate realistic, functional genomic sequences
Nov 18, 2024
[DALL-E]
Imagine an AI model so powerful it can read the genetic code of life, understand it, and even write it. Meet Evo, a groundbreaking artificial intelligence model that's set to revolutionize synthetic biology by decoding and designing DNA, RNA, and protein sequences at scales never before possible.
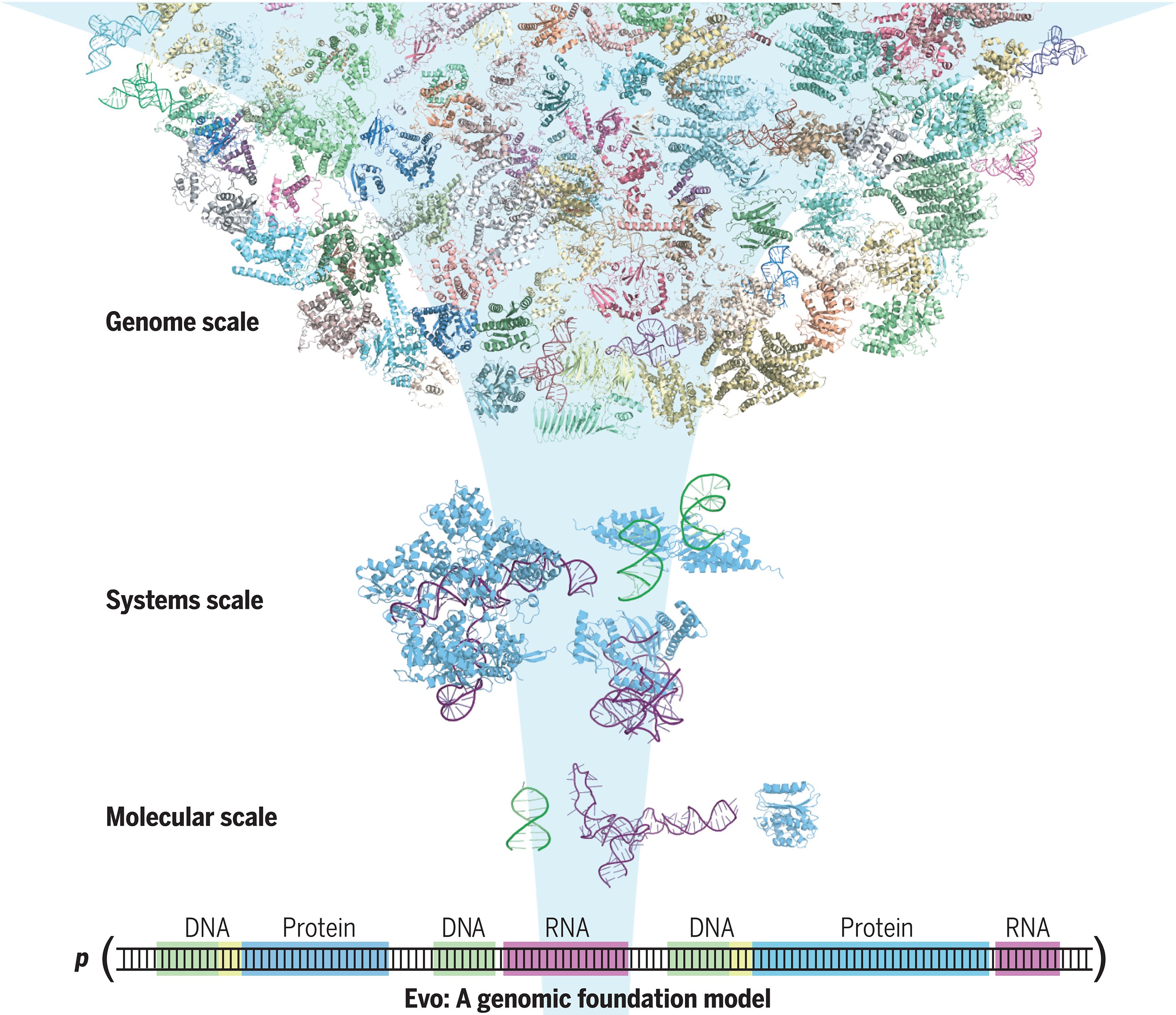
Developed by Eric Nguyen and his team, Evo is more than just another AI—it's a genomic foundation model equipped with seven billion parameters. Trained on a staggering dataset of 2.7 million microbial genomes, Evo leverages advanced deep-learning techniques to process long DNA sequences with unprecedented efficiency. Data from the recently published study detailing the newly developed platform was published recently in Science.
"By analyzing millions of microbial genomes, Evo has developed a comprehensive understanding of life’s complex genetic code, from individual DNA bases to entire genomes," notes Di Jiang in an editor’s summary. This profound understanding enables Evo to predict how tiny changes in DNA can impact an organism's fitness, generate realistic genome-length sequences, and even design new biological systems. In fact, the model's capabilities have been validated in the lab, successfully creating synthetic CRISPR systems and IS200/IS605 transposons.
Cracking the Code of Life
DNA, with its four-letter nucleotide vocabulary, encodes all genetic information essential for life. Variations in these sequences reflect adaptations honed by evolution over millions of years, allowing organisms to thrive in changing environments. Advances in DNA sequencing have mapped these variations across entire genomes, offering a treasure trove of data. But making sense of this vast information has been a monumental challenge.
Previous attempts to model DNA using techniques inspired by large language models (LLMs) have fallen short. These models often focused narrowly on individual molecules or segments of DNA, missing the bigger picture of genomic interactions crucial for understanding complex biological processes. Computational limitations further constrained their scope.
That's where Evo steps in.
A New Era of Biological Understanding
Evo isn't just larger; it's smarter. Built on the StripedHyena architecture, it's designed to handle sequences up to whole-genome scale. According to Nguyen and colleagues, Evo excels in both predictive and generative tasks. It achieves high accuracy in zero-shot evaluations for predicting mutation impacts on bacterial proteins and RNA, as well as in modeling gene regulation.
Perhaps most impressively, Evo understands the intricate dance between coding and noncoding sequences—the coevolution that defines complex biological systems. This enables the model to design functional CRISPR-Cas molecular complexes and transposable elements, marking the first time a language model has been used for protein-RNA and protein-DNA codesign.
"The genome is a sequence that encodes the DNA, RNA, and proteins that orchestrate an organism’s function," the researchers write. "We present Evo, a long-context genomic foundation model with a frontier architecture trained on millions of prokaryotic and phage genomes."
At the genomic scale, Evo can generate sequences over one megabase in length, vastly surpassing prior models. This capability opens doors to creating entire synthetic genomes with plausible architecture, a feat that was science fiction just a few years ago.
Implications for Medicine and Biotechnology
The potential applications of Evo are staggering. "The ability to predict the effects of mutations across all layers of regulation in the cell and to design DNA sequences to manipulate cell function would have tremendous diagnostic and therapeutic implications for disease," writes Christina Theodoris in a related perspective.
Consider personalized medicine: With Evo's predictive power, we could foresee how genetic mutations might influence an individual's health, leading to tailored treatments. In biotechnology, designing organisms with specific functions—like bacteria that consume pollutants or produce biofuels—becomes more feasible.
Moreover, Evo's approach could accelerate vaccine development. By understanding and predicting how viruses mutate, we could stay one step ahead in combating infectious diseases.
The Road Ahead
While Evo is a monumental leap forward, it's just the beginning. The researchers acknowledge that there's more to explore. "Future models may learn from diverse human and other eukaryotic genomes, using larger context lengths to capture distant genomic interactions over larger genomic scales," Theodoris suggests.
This means that as computational power grows and algorithms improve, models like Evo could eventually tackle the even more complex genomes of plants, animals, and humans. The implications for understanding diseases like cancer, which involve numerous genetic interactions, are profound.
Bridging Biology and Technology
Evo represents a convergence of biology and artificial intelligence, showcasing how machine learning can unlock secrets hidden in our genetic code. By capturing the multimodality of the central dogma—DNA, RNA, and proteins—and the multiscale nature of evolution, Evo enables prediction and design tasks from the molecular level up to entire genomes.
"Evo learns both of these representations from the whole-genome sequences of millions of organisms to enable prediction and design tasks from the molecular to genome-scale," the researchers explain. "Further development of large-scale biological sequence models like Evo, combined with advances in DNA synthesis and genome engineering, will accelerate our ability to engineer life."
A New Frontier in Synthetic Biology
Evo's emergence signals a new frontier where the limits of synthetic biology are redefined. It's a tool that doesn't just read the code of life—it writes it.
As Walter Beckwith aptly summarizes, "Evo’s ability to predict, generate, and engineer entire genomic sequences could change the way synthetic biology is done."
The journey ahead is exciting and uncharted. With models like Evo, we're not just observers of evolution—we're becoming active participants, harnessing the power of life's code to shape the future.


















