AI Uncovers Novel Proteins for Base Editing
This new discovery, based on AI-assisted structural predictions, could greatly expand the utility of base editors for therapeutic and agricultural applications
Jun 27, 2023
Image by Canva
In a significant stride towards advancing the biotechnology field, investigators from the Chinese Academy of Sciences' Institute of Genetics and Developmental Biology have trailblazed an AI-assisted method to unearth novel deaminase proteins. Through structural prediction and classification, the team has unlocked new functionalities that could revolutionize both therapeutic and agricultural applications. The findings of this groundbreaking study were recently published in Cell.
The world of biotechnology has seen swift progress due to the discovery of new proteins and the exploitation of various engineered enzymes. However, the traditional method of mining for these novel proteins, which relies on amino acid sequences, often falls short of providing a solid connection between protein structure and function.
Caixia Gao, PhD, the principal investigator on this study, has made headway in overcoming this challenge through base editing, a precision genome editing technology that is revolutionizing molecular crop breeding by enabling the introduction of desirable traits into elite germplasm. Despite many proteins being identified through sequence-based efforts for base editing, the limitations in editing specific DNA sequences or species have always been a hurdle.
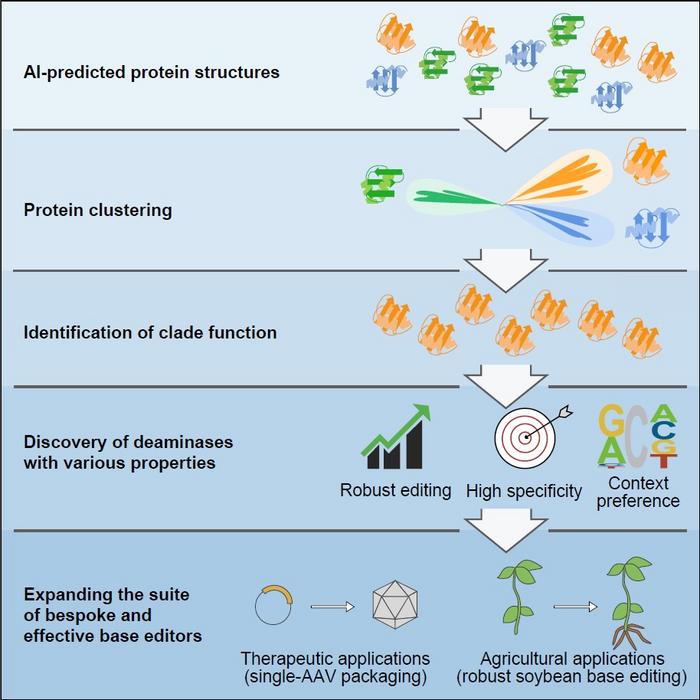
Historical efforts focused solely on protein engineering and directed evolution have diversified base editing properties, yet challenges endure. By employing the AI system, AlphaFold2 to predict the structures of proteins in the deaminase family, the researchers clustered and analyzed deaminases based on structural similarities. Their AI-assisted method led to the identification of five novel deaminase clusters exhibiting cytidine deamination activity, an essential function in the context of DNA base editors.
“We described the use of AlphaFold2 to predict and subsequently cluster an entire protein family based on predicted structure similarities. We selected deaminase proteins to analyze and identified many previously unknown properties,” the authors wrote. “We were surprised to find that most proteins in the DddA-like clade were not double-stranded DNA deaminases. We engineered the smallest single-strand-specific cytidine deaminase, enabling efficient cytosine base editor (CBE) to be packaged into a single adeno-associated virus (AAV). Importantly, we profiled a deaminase from this clade that edits robustly in soybean plants, which previously was inaccessible to CBEs.”
Through this innovative approach, the team reclassified a group of cytidine deaminases, SCP1.201, initially believed to act on dsDNA, and determined that it primarily performs deamination on ssDNA. With additional protein profiling and engineering, the researchers unveiled a suite of new DNA base editors boasting remarkable properties such as heightened efficiency, reduced off-target editing events, editing at varying preferred sequence motifs, and much smaller size.
The group emphasized that the development of this suite of base editors opens the door for future customized applications for medical therapies and agricultural breeding. Among the most notable achievements is the development of the smallest single-strand specific cytidine deaminase, which allowed for the efficient packaging of the first cytosine base editor into a single adeno-associated virus. Additionally, the team discovered a highly effective deaminase specifically for soybean plants, a crop of global significance that previously exhibited poor editing by cytosine base editors.
The researchers' findings underscore the transformative potential of AI-assisted structural predictions, particularly in the cytidine deaminase superfamily, to create a suite of innovative technologies and reveal new protein functions. These newly discovered deaminases have the potential to significantly broaden the range of base editors for therapeutic and agricultural applications.
This breakthrough research is anticipated to spark broad interest within the larger scientific community, particularly those in fields such as phylogenetics, metagenomics, protein engineering and evolution, genome editing, and plant breeding. By leveraging growing genomic databases for protein structure prediction, the pace of developing novel bioengineering tools is expected to accelerate.


















